A tutorial on investigate genetic admixture using STRUCTURE software
Updated:
Structure Software is a freely available software package that one may use for rigorous investigation of admixed individuals; identification of point of hybridization and migrants; and estimate over all structure of a population using a commonly used genetic markers such as SNPs and SSRs. This software was developed by Pritchard Lab at Stanford University and can downloaded at this link.
Download sample data set: click here
In this tutorial, I will show how to prepare input files and run the Structure software. For detail information, please read this article at this link
Step 1: Preparing the Input file
In this tutorial, I am using numerical SNP data as in input genotype file. One can convert their genotype data in numerical format in TASSEL software or any software package available as per ones convenience. The file needs to be formatted properly as shown below in the image below and save it as .txt file.
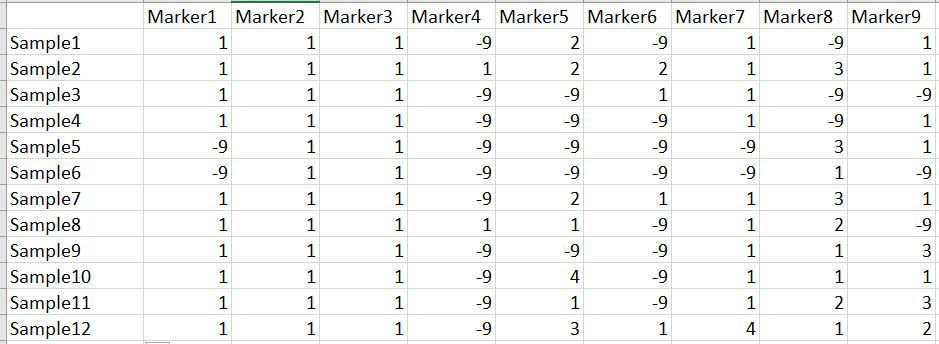
Please Note Missing data is denoted as -9 in the above image.
Step 2: Running the Structure biostat
1.1 Importing input file
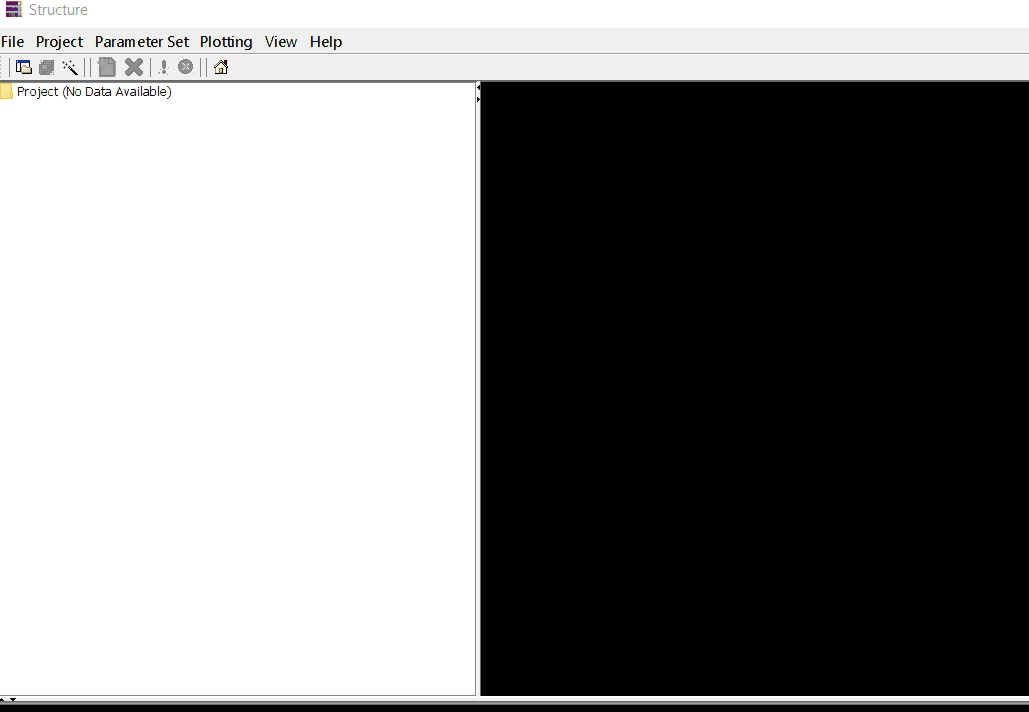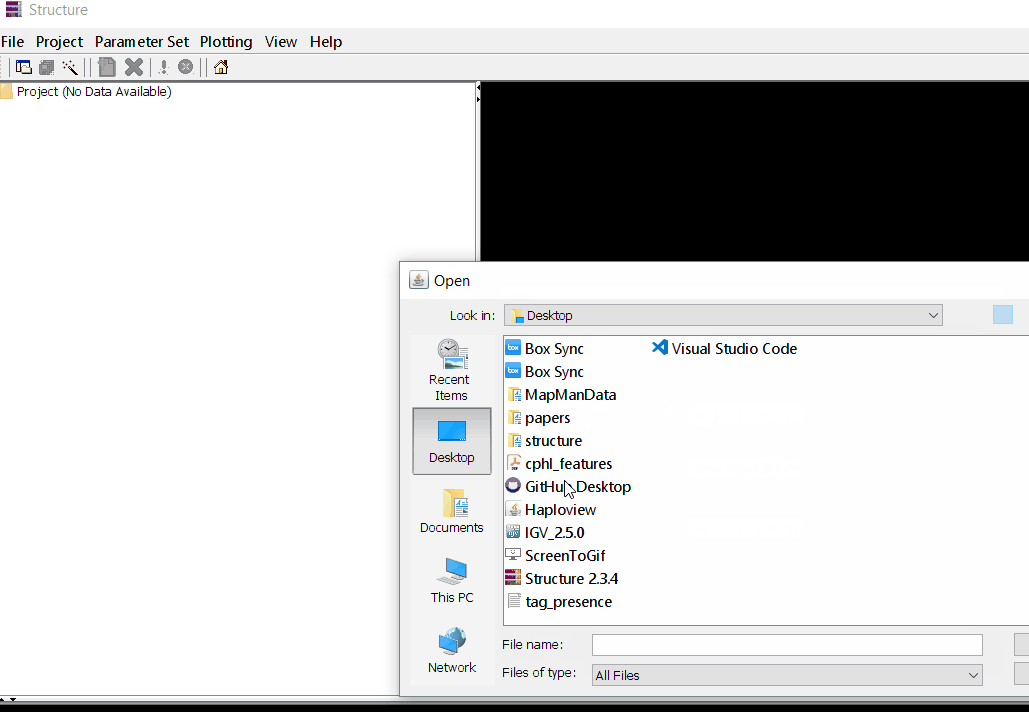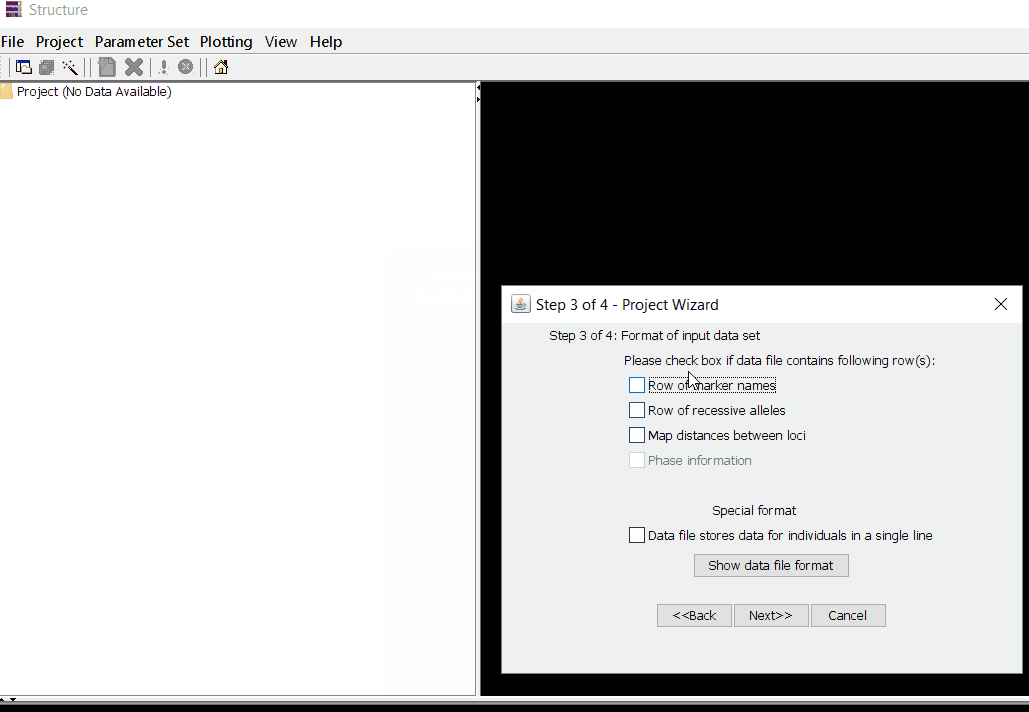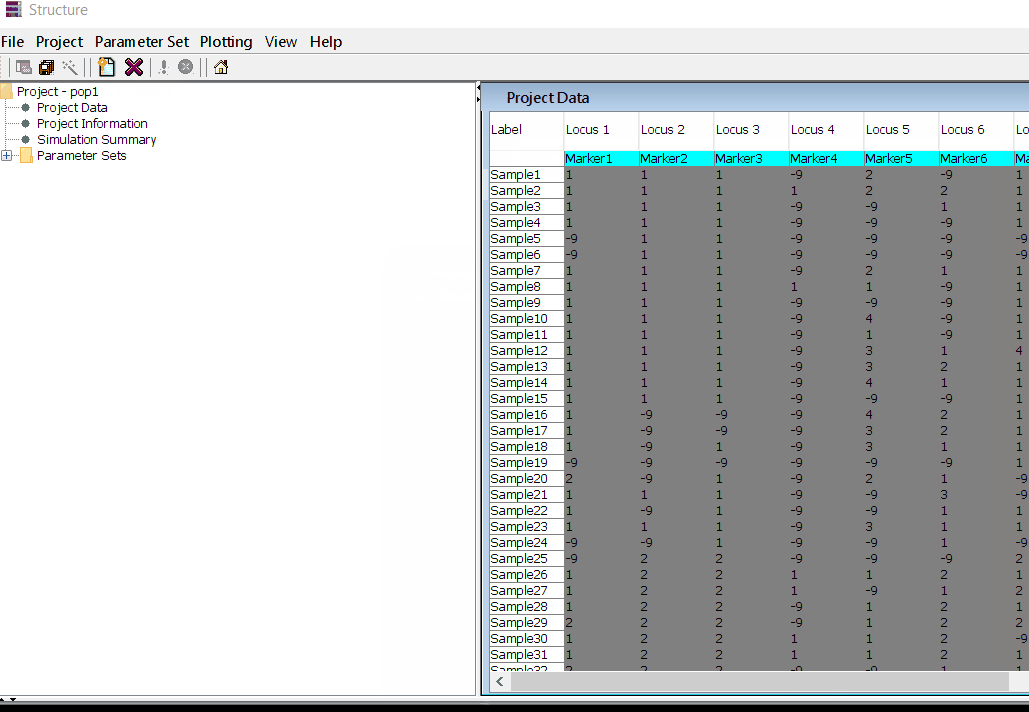
Once the input file with the correct header and format is ready, import the the file in Structure software using the steps shown in the below figure. The importing steps include 4 steps, please make sure to select correct directory and file name. At step 2 of 4 make sure to correctly input number of markers, samples/individual, and ploidy (if genotypes are ‘A’ then enter 1 but if, it is ‘AA’ enter 2), and finally, enter how the missing data are indicated as in the file. In this tutorial, I denoting the missing data ‘-9’.
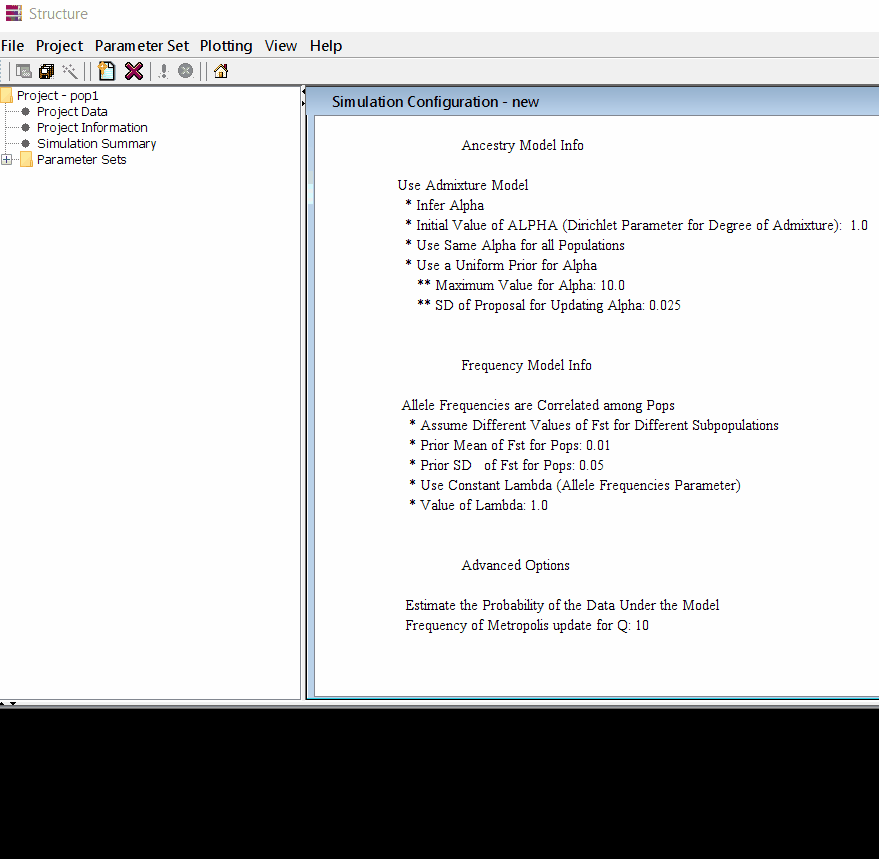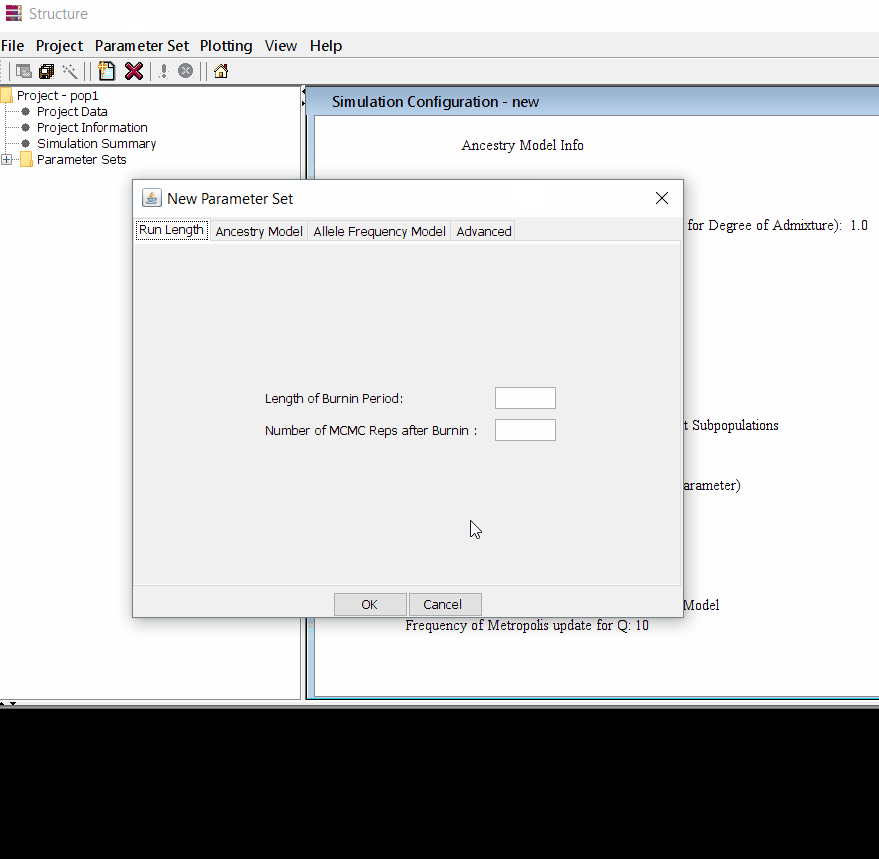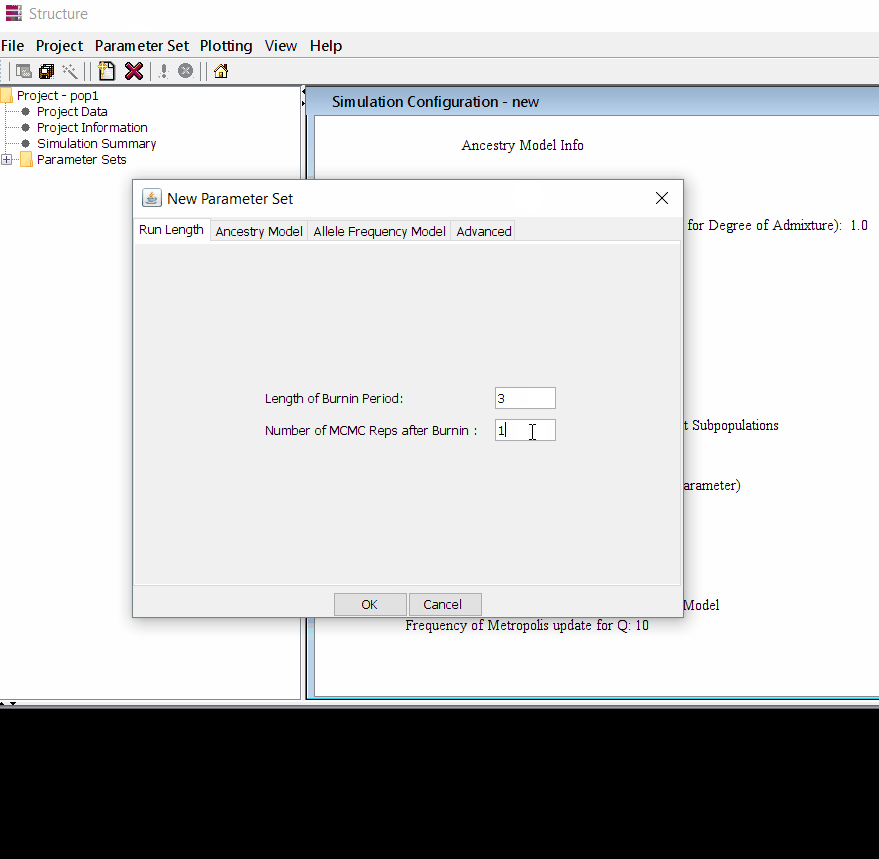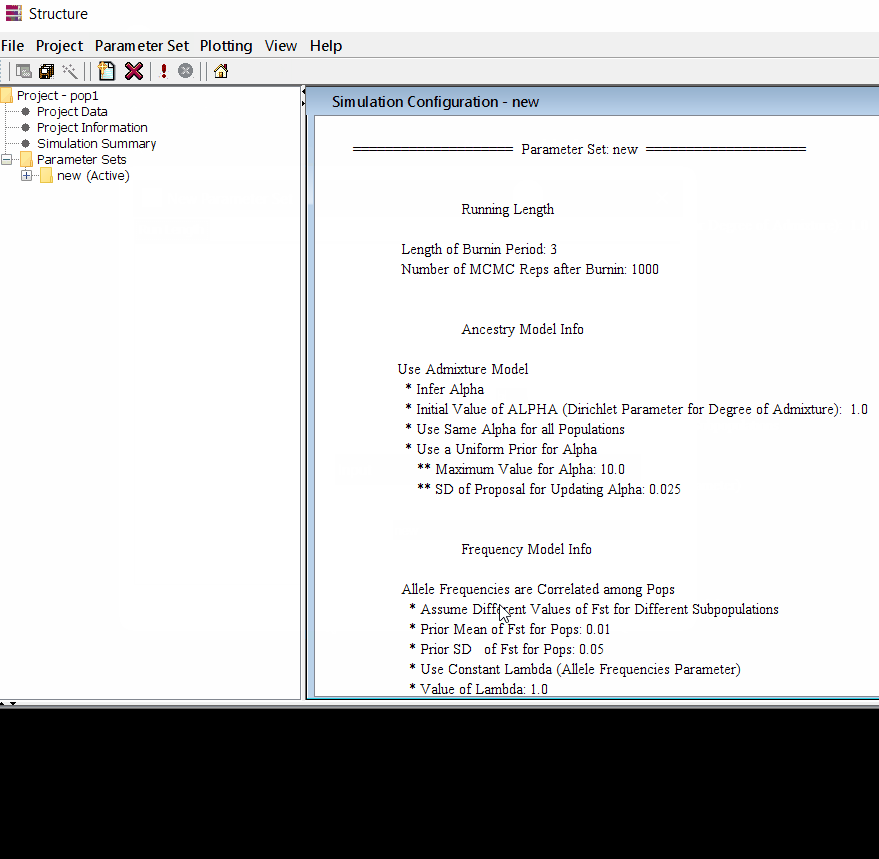
Step: 1.2 Set Parameter
Follow the steps shown in the below figure to run this step. Please remember One make custom add the length of burning period and Number of MCMC Reps after burnin.
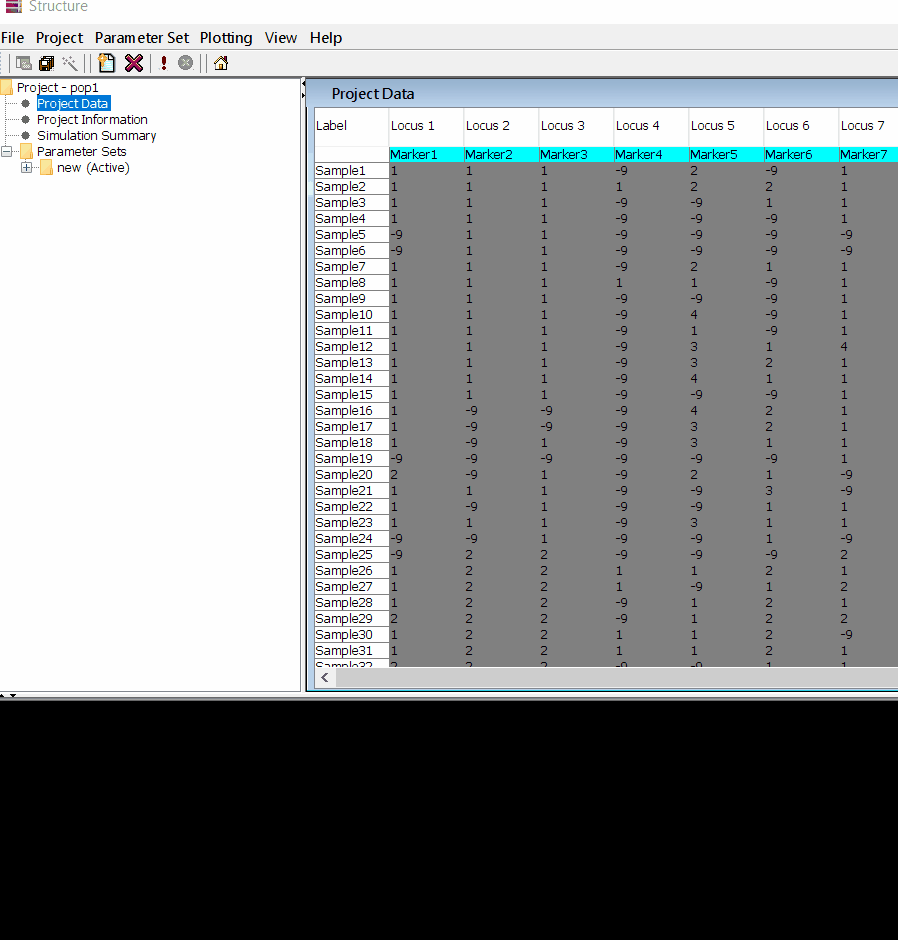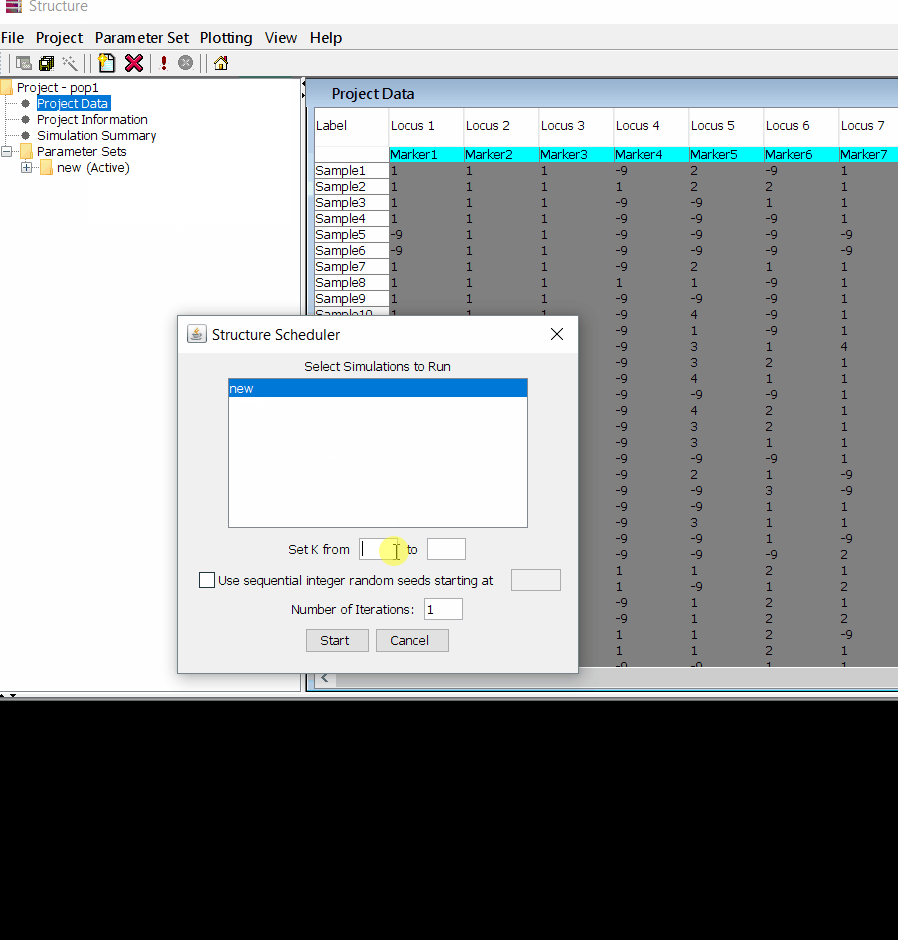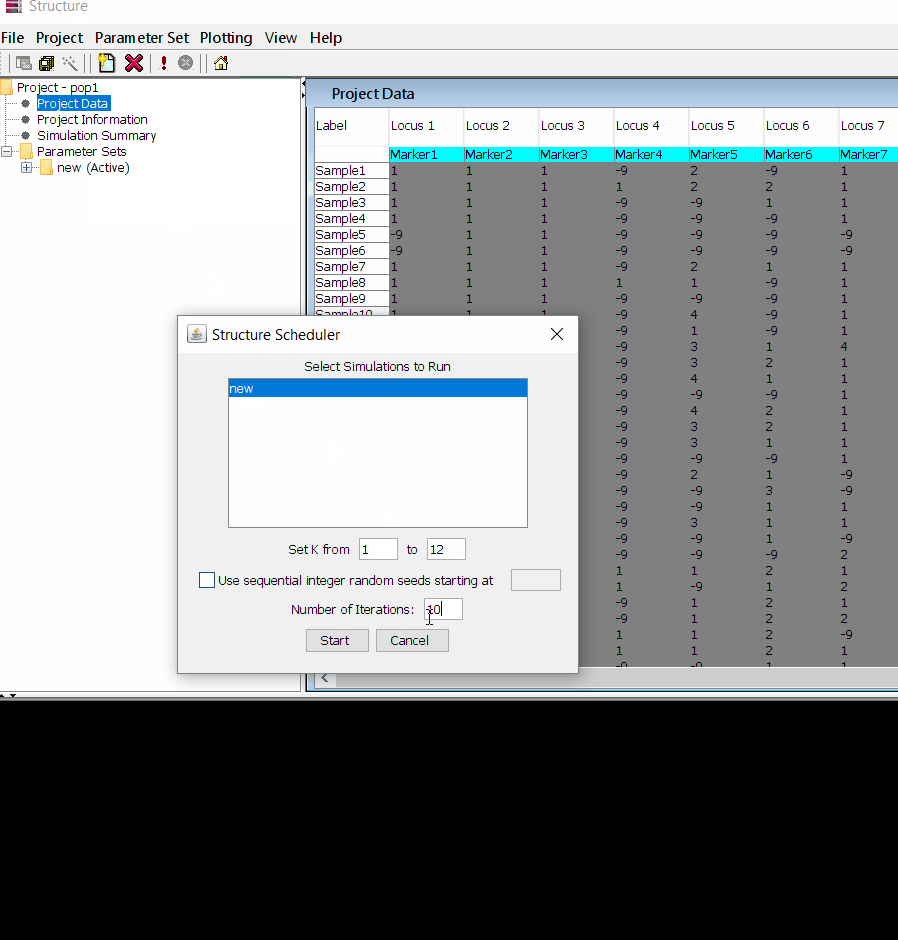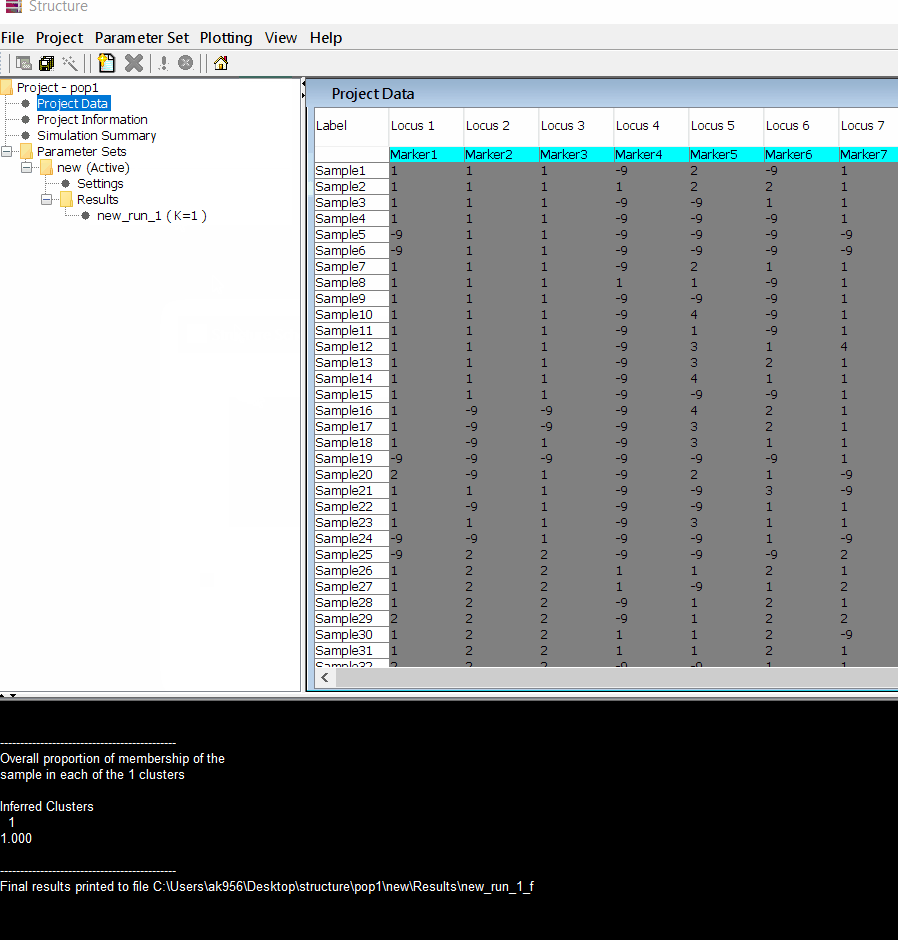
Step: 1.3 Running the project
Follow the steps shown in the below figure to run this step. Please remember to run at least 10 number of iterations. One see the job progress at the bottom black window of the shell.
Step: 1.4 Viewing the results
Follow the steps shown in the below figure to run this step. Please remember under the Results folder there are several branches of the results with various k values, which indicates number of sub-populations estimated from the given genetic data. It can tricky to pick the correct number of k for the data, and to solve this follow the next step to prepare files for a different software known as Structure Harvester.
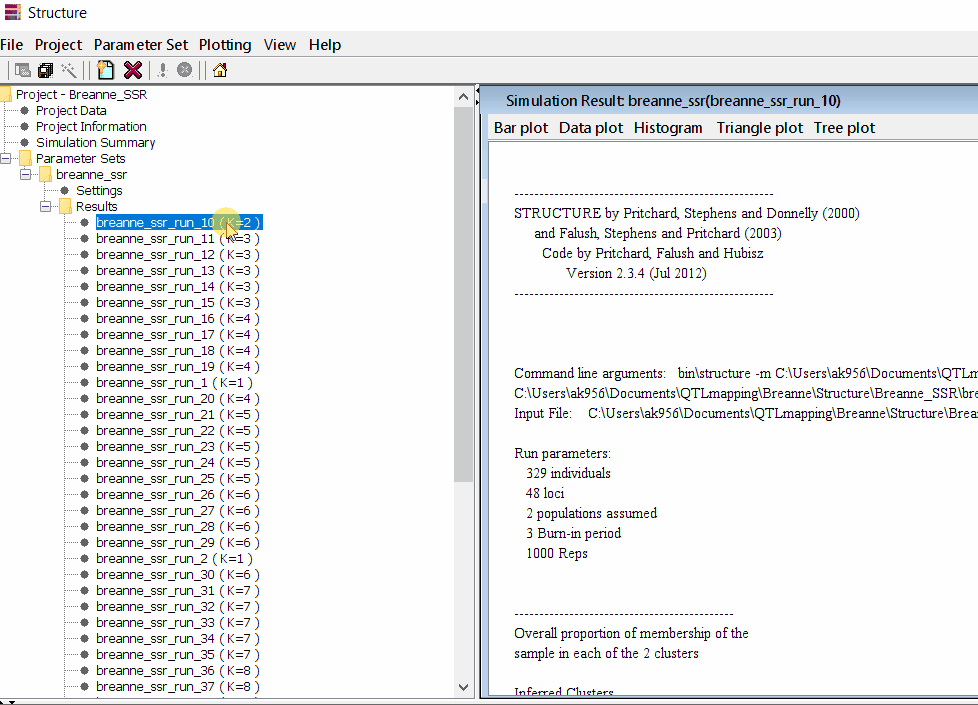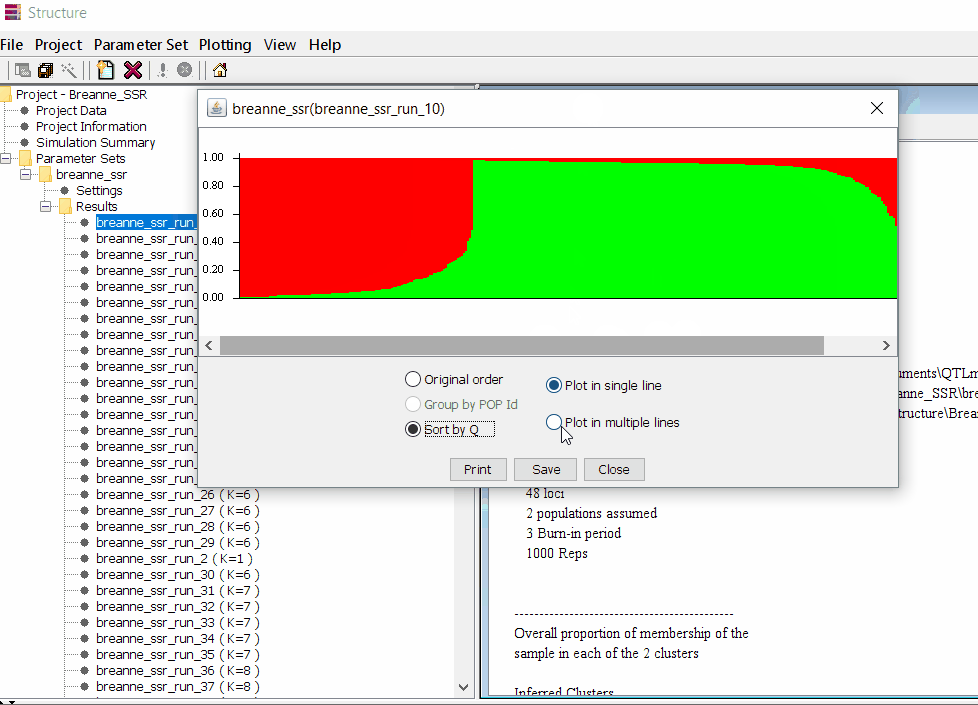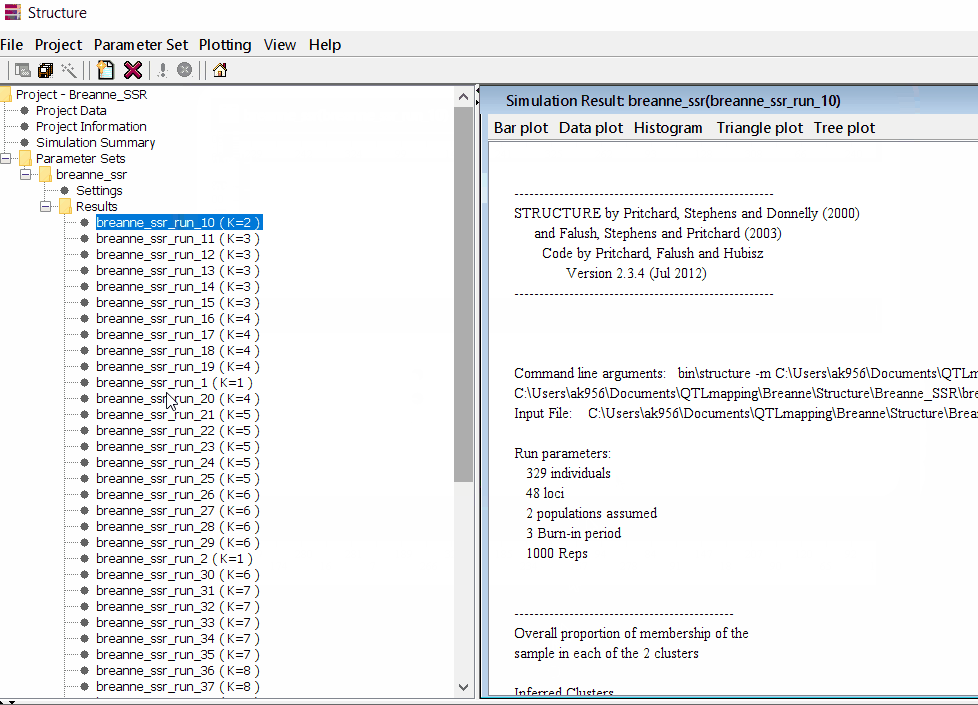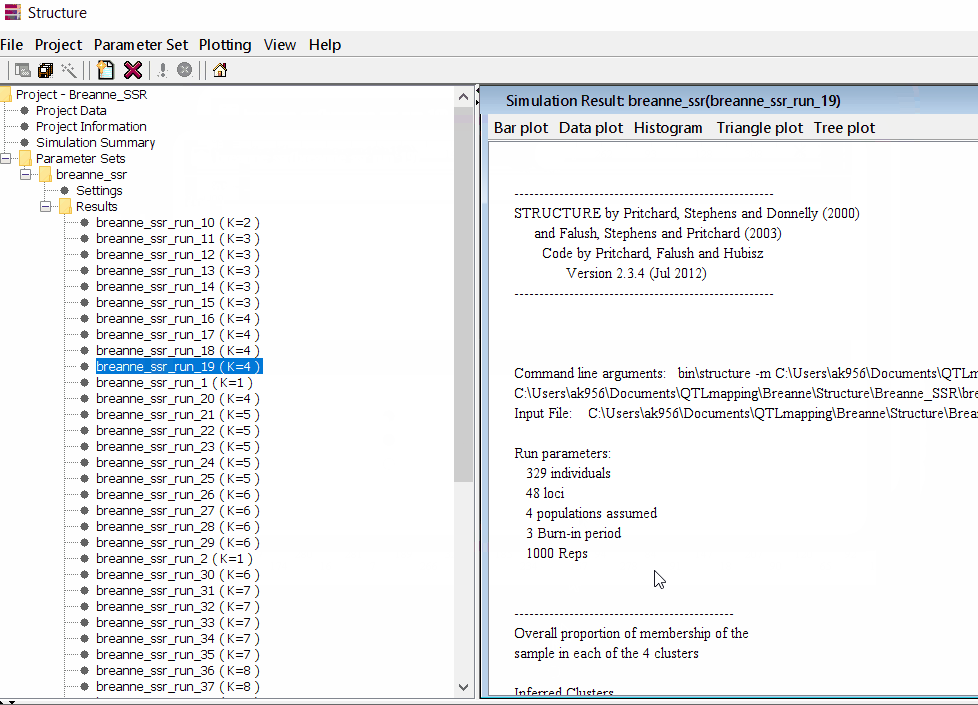
2.1 Preparing Files for Structure Harvester
zip all the result files in the results folder.
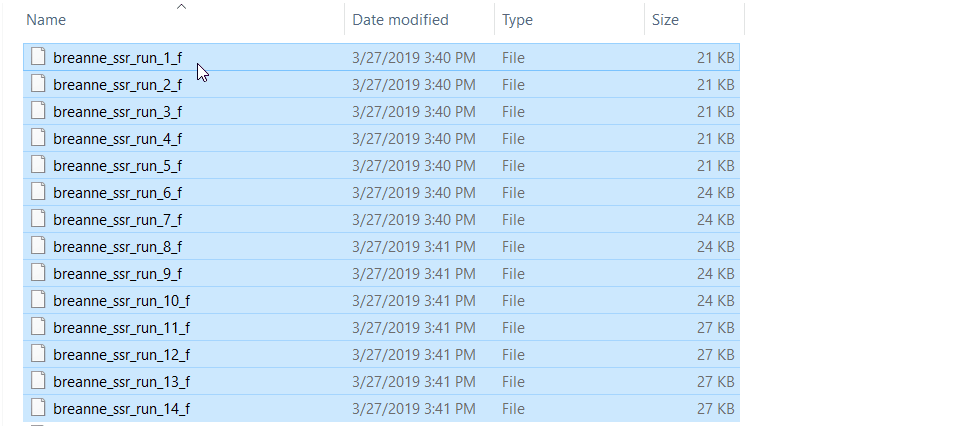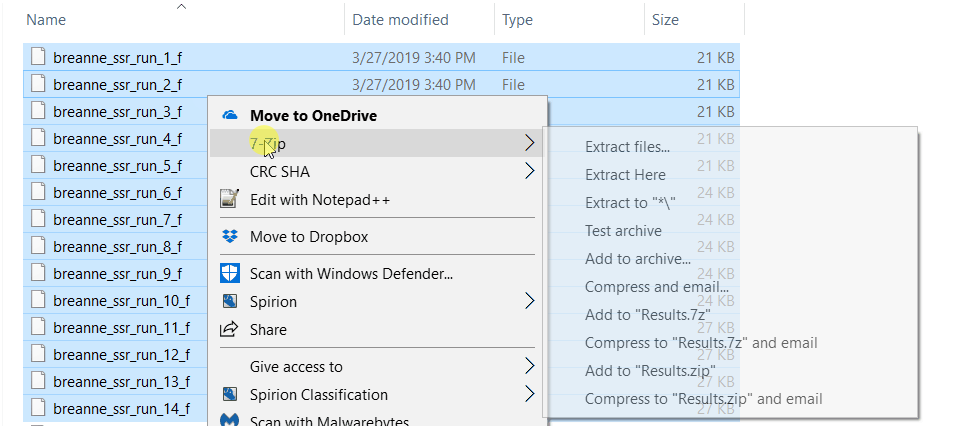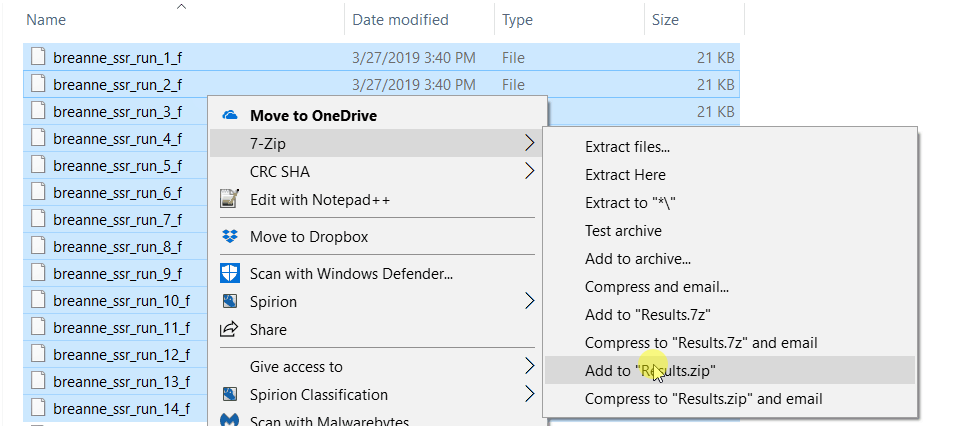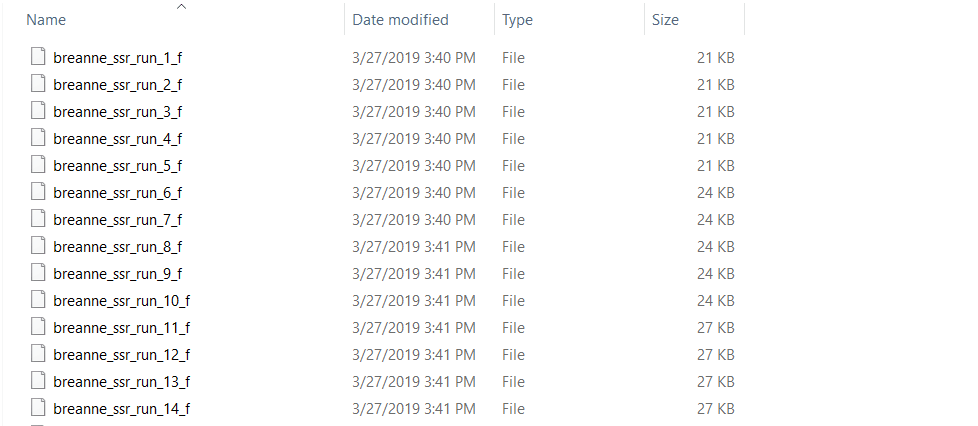
2.2 Running Structure Harvester
One your web browser search for structure harvester, and click the first the search result. Next, upload the results.zip file, click harvest to run the Structure Harvester program. It can take about few mins to run, however, it definitely depends on your data. Once the job is completed, the program outputs the summary of the analysis, the key output to look at is Delta K plot and Evanno table.
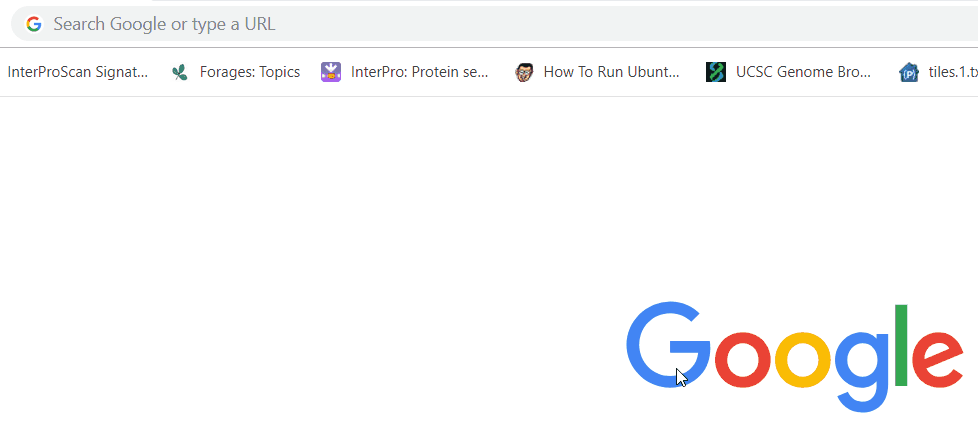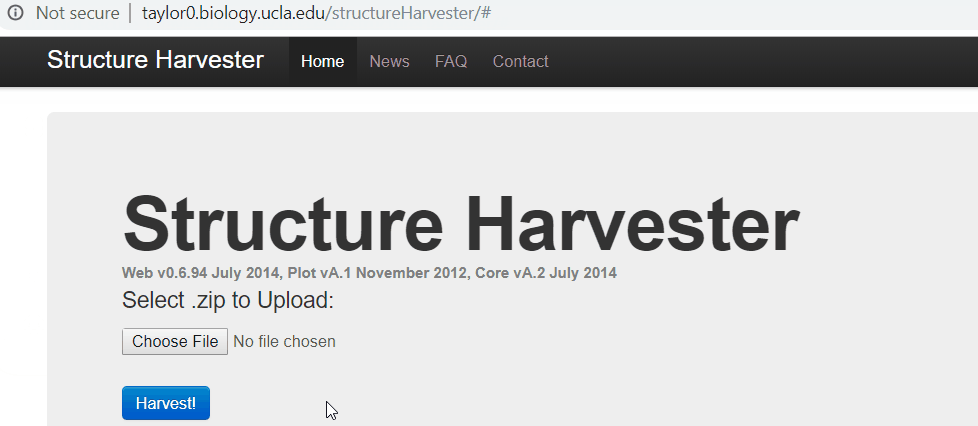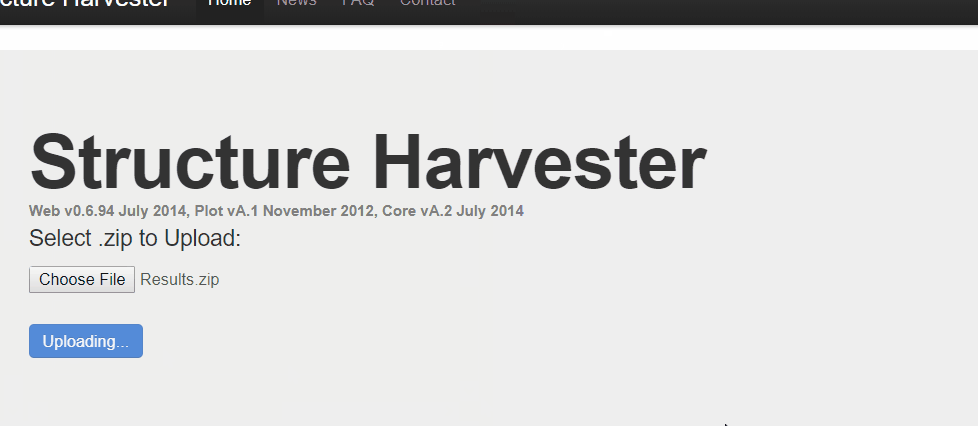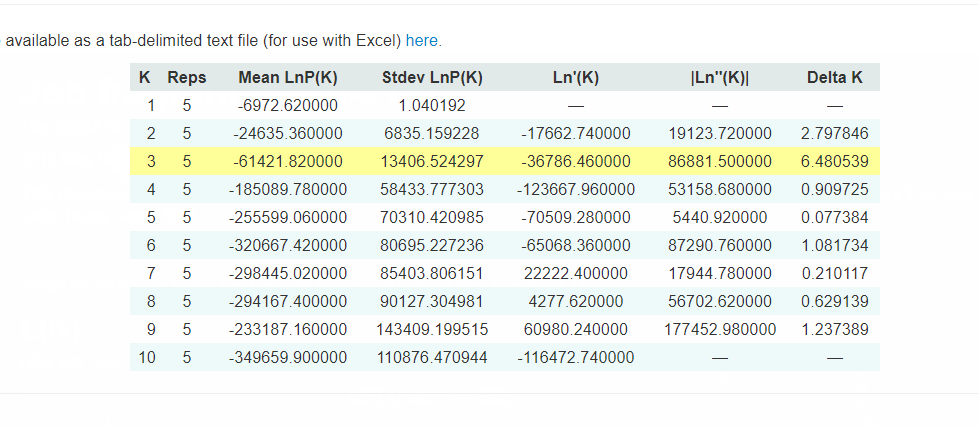
2.3 Interpreting the output
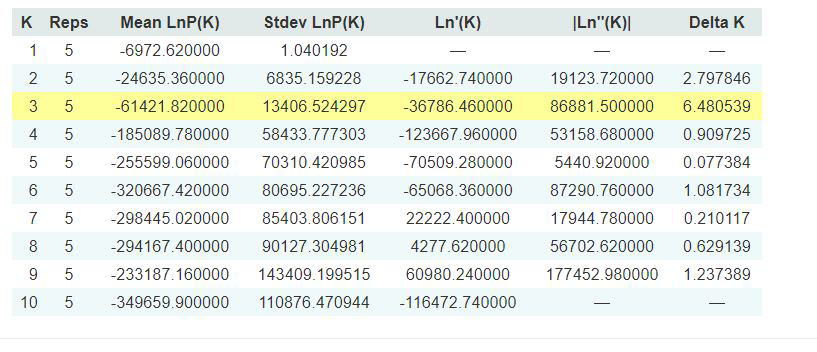
Evanno table highlights the significant k value that is estimated for this genotype data (see below figure). For this tutorial data set, the estimated from k is 3 subpopulation which is also supported by the Delta K plot, where a clear peak is see at K = 3 (see Delta K plot below).
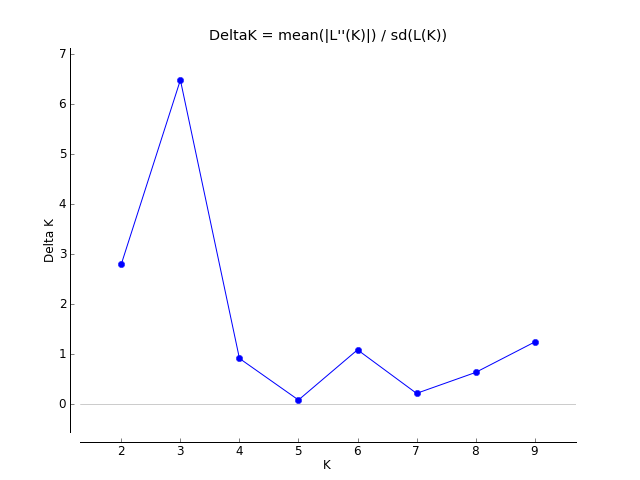
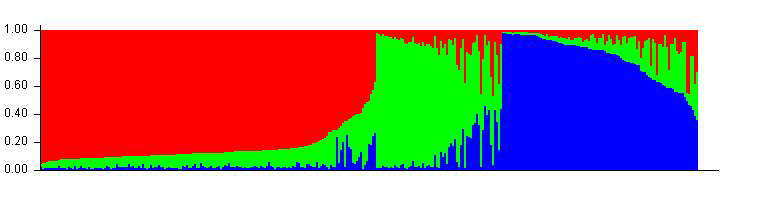
Therefore the correct bar plot with correct number of sub-population (k) is 3, which can be plotted by following the steps shown in 1.4
Thank you for reading this tutorial. If you have any questions or comments, please let me know by email.
Happy Structure-ing ![]()
Bibliography
Pritchard, Jonathan K., William Wen, and Daniel Falush. "Documentation for STRUCTURE software: Version 2." (2003).
Earl, Dent A. "STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method." Conservation genetics resources 4.2 (2012): 359-361.

Leave a comment